Software

ANKAphase
ANKAphase processes X-ray inline phase-contrast radiographs by reconstructing the projected thickness of the object(s) imaged. The tool uses a single-distance non-iterative phase-retrieval algorithm described in a paper by D. Paganin et al. J. Microsc. vol. 206 (2002). It has an easy-to-use graphical user interface and can be run either as a standalone application or as a plugin to ImageJ. It works on powerful clusters but also on your office laptop.
Atomic Simulation Environment (ASE)
The Atomic Simulation Environment (ASE) is a set of tools and Python modules for setting up, manipulating, running, visualizing and analyzing atomistic simulations.

ATSAS
ATSAS is a program suite for small-angle scattering data analysis from biological macromolecules. It includes multiplatform data manipulation and displays tools, programs for automated data processing and calculation of overall parameters, usage of high- and low-resolution models from other structural methods, algorithms to build three-dimensional models from weakly interacting oligomeric systems and complexes, and enhanced tools to analyse data from mixtures and flexible systems.

Avizo
3D analysis software for scientific and industrial data. Different application areas: Materials Science, Quality Assurance in Industrial Environments, Electronics, Digital Rock Analysis etc. It enables users to perform interactive visualization and computation on 3D data sets. The Avizo interface is modelled on the visual programming. Users manipulate data and module components, organized in an interactive graph representation (called Pool), or in a Tree view. Data and modules can be interactively connected together, and controlled with several parameters, creating a visual processing network whose output is displayed in a 3D viewer. With this interface, complex data can be interactively explored and analyzed by applying a controlled sequence of computation and display processes resulting in a meaningful visual representation and associated derived data.
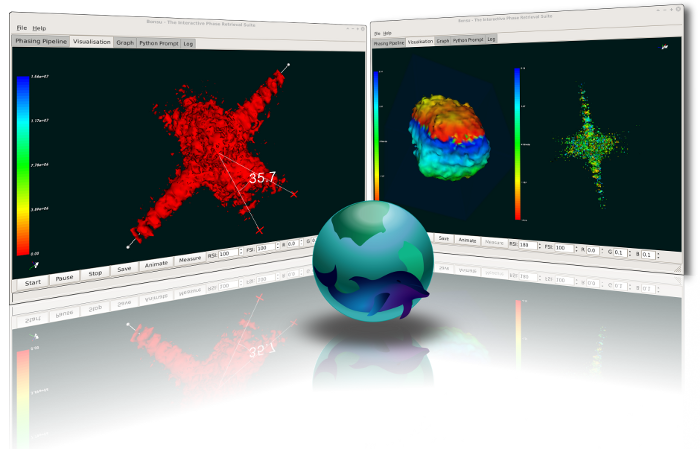
BONSU
Bonsu is an interactive phase retrieval suite, designed for phase retrieval with real-time visualisation in both two and three dimensions. It includes an inventory of algorithms and routines for data manipulation and reconstruction.
BornAgain
BornAgain is a software package to simulate and fit small-angle scattering at grazing incidence. It supports analysis of both X-ray (GISAXS) and neutron (GISANS) data. Its name, BornAgain, indicates the central role of the distorted wave Born approximation in the physical description of the scattering process. The software provides a generic framework for modeling multilayer samples with smooth or rough interfaces and with various types of embedded nanoparticles.
CCP4
CCP4 is an integrated suite of programs that allows researchers to determine macromolecular structures by X-ray crystallography, and other biophysical techniques. CCP4 aims to support the experimental determination and analysis of protein structures.
Cosmos (Lamp)
Data reduction utility to treat TOF reflectivity data. It is distributed with LAMP, but it was developed originally as a standalone IDL application and it can still be compiled and used without LAMP.

Crispy
Crispy is a modern graphical user interface to simulate core-level spectra using semi-empirical multiplet approaches.

CrysAlis Pro
Data collection and data processing software for small molecule and protein crystallography. Main features: automatic crystal screening, data collection and strategy modules. It provides the user with information regarding the unit cell, intensity estimation by resolution range and suggested frame exposure times for the full data collection.

CSD (CCDC)
The Cambridge Structural Database (CSD) is both a repository and a resource for the three-dimensional structural data of molecules generally containing at least carbon and hydrogen, comprising a wide range of organic, metal-organic and organometallic molecules. The specific entries are complementary to the other crystallographic databases such as the PDB, ICSD and PDF. The data, typically obtained by X-ray crystallography and less frequently by neutron diffraction, and submitted by crystallographers and chemists from around the world, are freely accessible (as deposited by authors) on the Internet via the CSD's parent organization's website (CCDC). ConQuest is the primary program for searching and retrieving information from the CSD.

DaFy
DaFy, short for Data Analysis Factory, is a software package that contains a bunch of PyQt5 based GUI applications for processing synchrotron X-ray data.

DAWN
DAWN, the Data Analysis WorkbeNch, is an Eclipse based application for scientific data analysis. It comes with a range of tools for visualization (1D, 2D and 3D), code development environments (for Python, Jython and Eclipse plug-ins) as well as processing workflows with visual algorithms for analyzing scientific datasets. It is primarily developed at Diamond Light Source, but external contributions are most welcome! DAWN is distributed freely and is released under the Eclipse Public License.

Demeter
Demeter is a comprehensive system for processing and analyzing X-ray Absorption Spectroscopy data. It contains several packages such as Athena, Artemis and Hephaestus, which are widely used in the XAFS community.

DiffPy-CMI
DiffPy-CMI is a library of Python modules for robust modeling of nanostructures in crystals, nanomaterials, and amorphous materials. The software provides functionality for storage and manipulation of structure data and calculation of structure-based quantities, such as PDF, SAS, bond valence sums, atom overlaps, bond lengths, and coordinations. Most importantly the DiffPy-CMI package contains a fitting framework for combining multiple experimental inputs in a single optimization problem.

