Software

ATSAS
ATSAS is a program suite for small-angle scattering data analysis from biological macromolecules. It includes multiplatform data manipulation and displays tools, programs for automated data processing and calculation of overall parameters, usage of high- and low-resolution models from other structural methods, algorithms to build three-dimensional models from weakly interacting oligomeric systems and complexes, and enhanced tools to analyse data from mixtures and flexible systems.
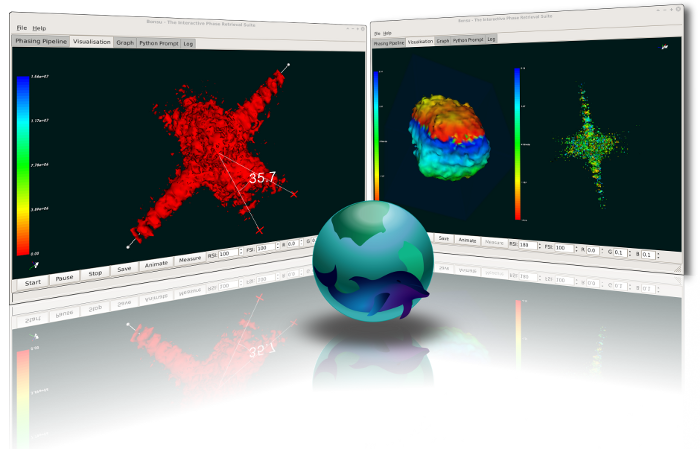
BONSU
Bonsu is an interactive phase retrieval suite, designed for phase retrieval with real-time visualisation in both two and three dimensions. It includes an inventory of algorithms and routines for data manipulation and reconstruction.
CCP4
CCP4 is an integrated suite of programs that allows researchers to determine macromolecular structures by X-ray crystallography, and other biophysical techniques. CCP4 aims to support the experimental determination and analysis of protein structures.

CrystFEL
CrystFEL is a suite of programs for processing diffraction data acquired "serially" in a "snapshot" manner, such as when using the technique of Serial Femtosecond Crystallography (SFX) with a free-electron laser source. CrystFEL comprises programs for indexing and integrating diffraction patterns, scaling and merging intensities, simulating patterns, calculating figures of merit for the data and visualising the results. Supporting scripts are provided to help at all stages, including importing data into CCP4 for further processing. [From: the Website]
FIT2D
FIT2D is a general purpose 1 and 2 dimensional data analysis program. It is used for both interactive and "batch" data processing, and is used for different purposes. Calibration and correction of detector distortions is one of the main uses of FIT2D. Difficult data analysis problems may be tackled using fitting of user specified models. To enable model fitting to be performed on a wide variety of input data, many other more basic data analysis operations are also available. A wide variety of performant graphical display methods are available.
GROMACS
GROMACS (GROningen MAchine for Chemical Simulations) is a molecular dynamics and energy minimization package mainly designed to simulate the Newtonian equations of motion for systems with hundreds to millions of particles, typically of proteins, lipids and nucleic acids. GROMACS can run on CPUs and GPUs.
iFit
The iFit library (pronounce [eye-fit]) is a set of methods to load, analyse, plot, fit and optimize models, and export results. iFit is based on Matlab, but can also be launched without Matlab license (stand-alone version).Matlab It does not currently include advanced graphical user interfaces (GUI), and rather focuses on doing the math right. Any text file can be imported straight away, and a set of binary files are supported. Any data dimensionality can be handled, including event based data sets (even though not all methods do work for these). Any model can be assembled for fitting data sets. Last, a number of routines are dedicated to the analyses of S(q,w) and S(alpha,beta). More advanced features include the full automation to compute phonon dispersions in materials, using DFT codes such as ABINIT, ELK, VASP, QuantumEspresso, GPAW and more (Models/sqw_phonons). The software can also compute the neutron TAS resolution function (4D) and fits to experimental data with full resolution convolution (ResLibCal). An interface for McStas and McXtrace is also available to automate and optimize instrument simulations.

McStas
A neutron ray-trace simulation package. McStas is a general tool for simulating neutron scattering instruments and experiments.
McXtrace
Monte Carlo Xray Tracing. Allows simulation of X-ray beam lines, as well as sample simulations in so-called virtual experiments.
MOLDY
Moldy is a C program for performing molecular-dynamics simulations of solids and liquids using periodic boundary conditions. The model system is completely specified in a run-time input file and may contain atoms, molecules or ions in any mixture. Molecules or molecular ions are treated in the rigid-molecule approximation and their rotational motion is modeled using quaternion methods. The equations of motion are integrated using a modified form of the Beeman algorithm. Simulations may be performed in the usual NVE ensemble or in isobaric and/or isothermal ensembles. Potential functions of the Lennard–Jones, 6-exp and MCY forms are supported and the code is structured to give an straightforward interface to add a new functional form. The Ewald method is used to calculate long-ranged electrostatic forces.
Pore3D
Pore3D is a software toolbox for quantitative analysis of three-dimensional images. The core of Pore3D consists in a set of state-of-the-art functions and procedures for performing filtering, segmentation, skeletonization and quantitative analysis of three-dimensional data. Although three-dimensional data can be produced by several techniques (for instance: magnetic resonance, x-ray scattering or confocal microscopy), the library was developed and optimized for micro-CT (Computed Tomography) data. Pore3D features are available through the high-level scripting environment IDL. Pore3D has been tested with IDL 64-bit from versions 6.4 to 8.5.
PyFAI
pyFAI is an azimuthal integration library that tries to be fast (as fast as C and even more using OpenCL and GPU). It is based on histogramming of the 2theta/Q positions of each (center of) pixel weighted by the intensity of each pixel, but parallel version uses a SparseMatrix-DenseVector multiplication
PyHST2
Hybrid distributed code for high speed tomographic reconstruction with iterative reconstruction and a priori knowledge capabilities. PyHST2 (formerly known as PyHST) has been engineered to sustain the high data flow typical of the third generation synchrotron facilities (10 terabytes per experiment) by adopting a distributed and pipelined architecture. The code implements, beside a default filtered backprojection reconstruction, iterative reconstruction techniques with a-priori knowledge. The latter are used to improve the reconstruction quality or in order to reduce the required data volume and reach a given quality goal. The implemented a-priori knowledge techniques are based on the total variation penalisation and a new recently found convex functional which is based on overlapping patches.
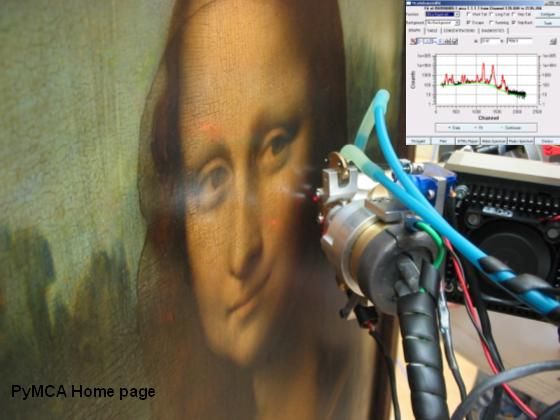
PyMca
X-ray Fluorescence Toolkit (visualization and analysis of energy-dispersive X-ray fluorescence data). . The program allows both interactive and batch processing of large data sets and is particularly well suited for X-ray imaging. Its implementation of a complete description of the M shell is particularly helpful for analysis of data collected at low energies. It features, among many other things, the fundamental parameters method
ROD
ROD is a program that can be used to do a refinement of a surface structure using surface X-ray diffraction data. All main features one encounters on surfaces, like roughness, relaxations, reconstructions and multiple domains, are taken into account. The most essential part of ROD is the calculation of the structure factor of the surface. ROD is complemented by two utilities: ANA and AVE: ANA can be used to integrate scans and to convert these into structure factors, while AVE can sort and average data, determine agreement factors and produce a data file for the program ROD.

